Category | Name | Core algorithms | Similarity measures | Databases | Numbersa | Cross-validation | Case study | Source code | Published |
|---|---|---|---|---|---|---|---|---|---|
Feature generation-based | GCNCDA [42] | FastGCN and Forest PA | Disease semantic similarity, GIP kernel similarity | CircR2Disease, MeSH | 739 circRNA–disease | 5-CV | Breast cancer, glioma, colorectal cancer | 2020.06 | |
GANCDA [43] | Generative adversarial network and logistic model tree | Disease semantic similarity, GIP kernel similarity | CircR2Disease, MeSH | 739 circRNA-disease | 5-CV | Gastric cancer, colorectal cancer, breast cancer | Unavailable | 2020.08 | |
DWNCPCDA [44] | DeepWalk, network consistency projection | CircRNA and disease topological similarity | CircR2Disease | 650 circRNA-disease | 5-CV | Hepatocellular carcinoma, lung cancer | 2021.02 | ||
NSL2CD [45] | DeepWalk and adaptive subspace learning | Disease semantic similarity, circRNA function similarity, GIP kernel similarity | CircR2Disease, MeSH | 649 circRNA–disease | 5-CV | Acute myeloid leukemia, lung cancer, breast cancer, etc. | Unavailable | 2021.05 | |
GATCDA [46] | Graph attention network | Disease symptom, network, entropy similarity, circRNA network entropy similarity, | CircR2Disease, CircAtlas 2.0, Circ2Disease, CircRNADisease, starBase v2.0, DisGeNET | 768 circRNA–disease | 5-CV | Bladder cancer, diabetes retinopathy, rheumatoid arthritis | Unavailable | 2021.06 | |
AE-RF [47] | Deep autoencoder, random forest | Disease semantic similarity, | CircR2Disease, DO | 650 circRNA–disease | 5-CV, | Breast cancer, colorectal cancer, lung cancer | 2020.11 | ||
IMS-CDA [48] | Stacked autoencoder, | Disease semantic similarity, disease and circRNA Jaccard similarity, GIP kernel similarity | CircR2Disease, MeSH | 739 circRNA–disease | 5-CV | Cardiovascular disease, glioma, intracranial aneurysms | 2021.11 | ||
Yang’s method [49] | accelerated attributed network embedding, stacked autoencoder, XGBoost | Disease semantic similarity, circRNA expression profile similarity, GIP kernel similarity | CircR2Disease, MeSH, | 605 circRNA–disease | 5-CV, 10-CV | Esophageal squamous cell carcinoma, gastric cancer | Unavailable | 2021.08 | |
Type discrimination-based | DeepDCR [50] | 24 meta-path-based, deep forest | Disease symptom similarity, miRNA functional similarity, circRNA co-expression similarity | Circ2Traits, miRTarBase, circBase, miRbase, GENCODE, TargetScan, miR2Disease, HMDD | 17,961 circRNAs, 469 miRNAs, and | 5-CV | Breast neoplasms, lung neoplasms, hepatocellular carcinoma, etc. | 2020.10 | |
Hybrid-based | MSFCNN [51] | Convolutional neural networks | Disease semantic, symptom, Lin, PSB, Resnik, SemFunSim similarity, circRNA sequence, regulatory, expression similarity, GIP kernel similarity, | CircR2Disease, CircBank, HMDD v3.0 | 325 circRNAs, 53 diseases and 3175 miRNAs | 5-CV | Acute myeloid leukemia, breast cancer, colorectal cancer, etc. | Unavailable | 2020.10 |
Wang’s method [52] | Convolutional neural network, extreme learning machine | Disease semantic similarity, GIP kernel similarity | CircR2Disease, MeSH | 739 circRNA–disease | 5-CV | Glioma, heart disease, cervical cancer, etc. | 2020.07 | ||
CDASOR [53] | Convolutional neural networks, recurrent neural networks | NA | CircR2Disease, Circ2Disease, circAtlas, LncRNAdisease, MNDR, circBase, UMLS, OMIM, circBase, DO | Dataset1: 754 circRNA–disease associations (630 circRNAs and 87 diseases), | 5-CV | Heart disease, colorectal cancer, hypoxia, etc. | 2021.05 | ||
AE-DNN [54] | Autoencoder, deep neural networks | Disease semantic similarity, circRNA sequence similarity, GIP kernel similarity | CircR2Disease, circBase | 445 circRNA–disease | 5-CV, 10-CV | Glioma, gastric cancer, liver cancer | Unavailable | 2020.10 | |
SGANRDA [55] | Generative adversarial network, extreme learning machine | Disease semantic similarity, circRNA sequence similarity, GIP kernel similarity | CircR2Disease, MeSH | 739 circRNA–disease | 5-CV, LOOCV | Cervical cancer, colorectal cancer, breast cancer, etc. | 2021.11 | ||
DMFCDA [56] | Deep matrix factorization, multi-layer neural networks | NA | CircR2Disease, | Dataset1: 619 circRNA–disease associations (556 circRNAs and 80 diseases), | 5-CV, LOOCV | Colorectal cancer, hepatocellular carcinoma, lung cancer, etc. | 2021.04 | ||
DMFMSF [57] | Weighted K-nearest known neighbor,singular value decomposition, deep matrix factorization | Disease semantic, hamming profile similarity, GIP kernel similarity | CircR2Disease, LncRNADisease, MeSH | Dataset1: 619 circRNA–disease associations (556 circRNAs and 80 diseases), | 5-CV, LOOCV | Hepatocellular carcinoma, breast cancer, acute myeloid leukemia | 2021.07 | ||
DMFCNNCD [58] | Deep matrix factorization, convolution neural network | Disease semantic similarity, circRNA function similarity | CircR2Disease, DO, | 650 circRNA–disease | 5-CV | Lung cancer, colorectal cancer, hepatocellular carcinoma | Unavailable | 2021.12 | |
Thosini’s method [59] | Graph convolution network | CircRNA sequence similarity, GIP kernel similarity | CircR2Disease, circBase | About 900 circRNA–disease | 5-CV | Stomach cancer | Unavailable | 2021.08 | |
CRPGCN [60] | Random walk with restart, principal component analysis, graph convolutional network | Disease semantic similarity, circRNA sequence, gene similarity, GIP kernel similarity | CircR2Disease, MeSH | 533 circRNAs and 89 | 2-CV, 5-CV, 10-CV | Breast cancer | 2021.11 | ||
RGCNCDA [61] | Random walk with restart, principal component analysis, relational graph convolutional networks | Disease semantic similarity, circRNA function similarity, GIP kernel similarity, miRNA expression similarity | CircR2Disease, HMDD v3.0, ENCORI, MeSH, | 650 circRNA–disease | 5-CV | Gastric cancer, coronary artery disease, bladder cancer, etc. | Unavailable | 2022.02 | |
IGNSCDA [62] | Graph convolutional network, multi-layer perceptron | CircRNA GIP kernel similarity, circRNA expression profile similarity | CircR2Disease, exoRBase | 612 circRNA–disease | 5-CV | Bladder cancer | Unavailable | 2021.09 | |
KGANCDA [63] | Graph attention network and Multiple layer perceptron | NA | circR2Cancer, lncRNASNP2, LncRNADisease, circad, circRNADisease | Dataset1: 514 circRNAs, 62 diseases, 564 miRNAs, 573 lncRNAs | 5-CV | Colorectal cancer | 2022.04 | ||
GMNN2CD [64] | Graph Markov neural network | Disease semantic similarity, GIP kernel similarity | CircR2Disease, Circ2Disease, circRNA–disease, circAtlas, CircFunBase, MeSH | Dataset1: 533 circRNA–disease associations (612 circRNAs and 89 diseases) | 5-CV | Breast cancer, cervical cancer | 2022.02 |
GCNCDA: A new method for predicting circRNA-disease associations based on Graph Convolutional Network Algorithm
https://github.com/look0012/GCNCDA/ 不可用
Materials and methods
Method overview
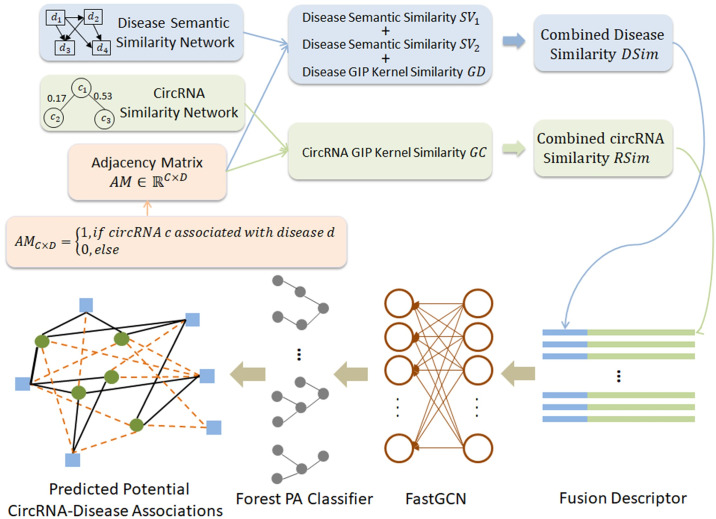
根据疾病语义相似性网络和circRNA-disease邻接矩阵构建疾病语义相似性矩阵和疾病高斯相互作用相似性矩阵(Gaussian interaction profile ,GIP)。
然后根据circRNA相似性网络和circRNA-disease邻接矩阵构建circRNA GIP相似性矩阵。3.
疾病和circRAN相似性矩阵融合。
FastGCN 提取高层特征,生成特征
ForestPA 分类器进行分类
Benchmark dataset
使用circR2Disease数据库作为benchmark,它包含661 circRNAs,100 diseases,和739 circRNA-disease联系。为了平衡,这里选择739 个负样本作为全部的负样本。
Construction of CircRNA similarity model
circRNA c(i) 的向量 V(c(i)) 为100维,表示和100个疾病的关系,相关设为1 否则为0。
circRNA c(i) 和circRNA c(j) 的GIP核心相似性 GC(c(i),c(j))
Construction of disease similarity model
疾病的GIP相似性构建 Gd(d(i),d(j)) 和 circRNA 构建方式一样
疾病的语义相似性用 MesSH数据库构建。它使用DAG反映不同疾病之间的关系。一个疾病d 机构用 DAG_d=(d,N_d,E_d) 表示 ,N_d表示所有和d有关系的节点,包括d ,E_d 表示这些病之间的关系。对于 DAG_d 内的疾病 s ,它的贡献值 D_d(s) 计算如下
这里是层级的关系,个人理解,d 值为1 所有和 d 上游(父节点)为 0.5 ,然后上游的上游为0.5*0.5,依次进行。
\mu 表示疾病和它的子疾病的贡献因子,这里取0.5。把所有疾病 d 相关疾病的值加起来得到它们的语义值
根据DAG中疾病的层级结构关系 疾病d(i) 和 d(j) 之间的语义相似性 SV_1(d(i),d(j))
DAG中不同疾病的数量也会影响其语义相似性
第二个语义相似性模型 SV_2(d(i),d(j))
实际计算后发现,由于661种circRNA和100种disease中只有很少一部分有关联,所以计算得到的矩阵十分稀疏,最后得到的相似度普遍过于接近,作用不大。
Multi-source data fusion
Rsim(i) 表示 Rsim矩阵中第 i 行,也就是 c(i) 和所有其他circRNA之间相似性形成的向量,DSim(j)同理。
Feature extraction by fast learning with Graph Convolutional Networks
Prediction by forest PA classifier
Inferring Potential CircRNA–Disease Associations via Deep Autoencoder‑Based Classification
https://github.com/Deepthi-K523/AE-RF
Materials and Methods
Datasets
CircR2Disease 数据库
相似性计算方面大差不差
Method
采用autoencoder和随机森林分类器去推理联系,一对circRNA disease 构建一个特征向量,所有的对构建的特征向量Q,Q输入到编码器中,获得一个低维的向量Q‘,再通过解码器尽量的还原成Q,然后用Q’获得的向量预测。
Autoencoder‑Based Feature Selection
Random Forest‑Based Association Prediction
RGCNCDA: Relational graph convolutional network improves circRNA-disease association prediction by incorporating microRNAs
Materials and methods
Data source and preprocessing
CircRNA-disease association
CircR2Disease
MiRNA-disease association
HMDD v3.0 有 32281条 miRNA-disease 其中 1206 miRNAs 和 893 diseases 过滤后选用了11,824条
CircRNA-miRNA interaction
ENCORI 中 2293条circRNA-miRNA 其中 68 of 13839 circRNAs and 581 of 642 miRNAs.
然后构建邻接矩阵 CD, CM, MD 代表关系
CD circRNA-disease associations
CM circRNA-miRNA interactions
MD miRNA-disease associations
Disease similarity network
Disease semantic similarity
同 1
Disease GAS similarity
同 1
Disease fusional similarity network
同 1
融合的相似性分数作为疾病相似性网络边的权重。
CircRNA similarity network
CircRNA functional similarity
疾病 d 和 疾病集合 D={d_1,d_2,...,d_n} 之间的语义相似度
这里 S_d 在上文中未出现过,文章的意思应该是上文中的 S_s .
circRNA c_i 和 circRNA c_j 之间的功能相似性
D_i,D_j 表示和 c_i, c_j 相关的疾病集合。
CircRNA GAS similarity
CircRNA fusional similarity network
Similarity calculation of miRNAs
计算基于这样的假设:具有相似表达模式的 miRNA 也具有相似的功能或生物学途径
miRNA i 和 miRNA j 之间的相似性可以通过miRNA表达谱之间的Pearson correlation coefficient (PCC) 计算
X, Y 作为 m_i,m_j 的基因表达谱,每个表达谱包括n列从不同类型的人类组织测量的表达值
Global heterogeneous network
RGCNCDA method
 重启随机游走算法(Random Walk with Restart,RWR)和 主成成分分析(principal component analysis PCA) 获取节点的特征输入到 RGCNCDA。然后使用R-GCN编码器和DistMult解码器构建一个预测模型。带有Laplacian regularization term 的损失函数计算最后的分数。
重启随机游走算法(Random Walk with Restart,RWR)和 主成成分分析(principal component analysis PCA) 获取节点的特征输入到 RGCNCDA。然后使用R-GCN编码器和DistMult解码器构建一个预测模型。带有Laplacian regularization term 的损失函数计算最后的分数。
relational graph convolutional networks (R-GCNs) 关系图神经网络
DistMult 模型用作衡量可信度的评分函数。
Experiments and results
一样的
Case study
GMNN2CD: identification of circRNA–disease associations based on variational inference and graph Markov neural networks
https://github.com/nmt315320/GMNN2CD
Materials and methods
 Datasets
Datasets
用了5个数据集 CircR2Disease , Circ2Disease , circRNA–disease , circAtlas and CircFunBase
Disease semantic similarity 1
同1
Disease semantic similarity 2
同1
Gaussian interaction profile kernel similarity for disease
同1
Gaussian interaction profile kernel similarity for circRNA
同1
Comprehensive similarity of multisource data fusion
同1
